2018
In the third annual Visualize This! competition participants could select their dataset: a bio-molecular dynamics simulation or a rich set of linked humanities data.
Interaction of a large protein structure with a cell’s membrane
The bio-molecular dynamics dataset was taken from a coarse-grained Gromacs simulation of interaction of a large protein structure with a cell’s membrane. A cell’s membrane is typically composed of two leaflets (single molecular layers), comprised in this model of pure phosphatidylcholine (POPC) lipid molecules. The protein structure is a P-glycoprotein (P-gp) embedded into the cell’s membrane. Such proteins are responsible for the translocation of a wide range of compounds across membranes.
A simple visualization with VMD (below) shows the horizontal cell membrane along with the embedded vertical protein. To further differentiate the two, we rendered the protein beads as larger spheres.
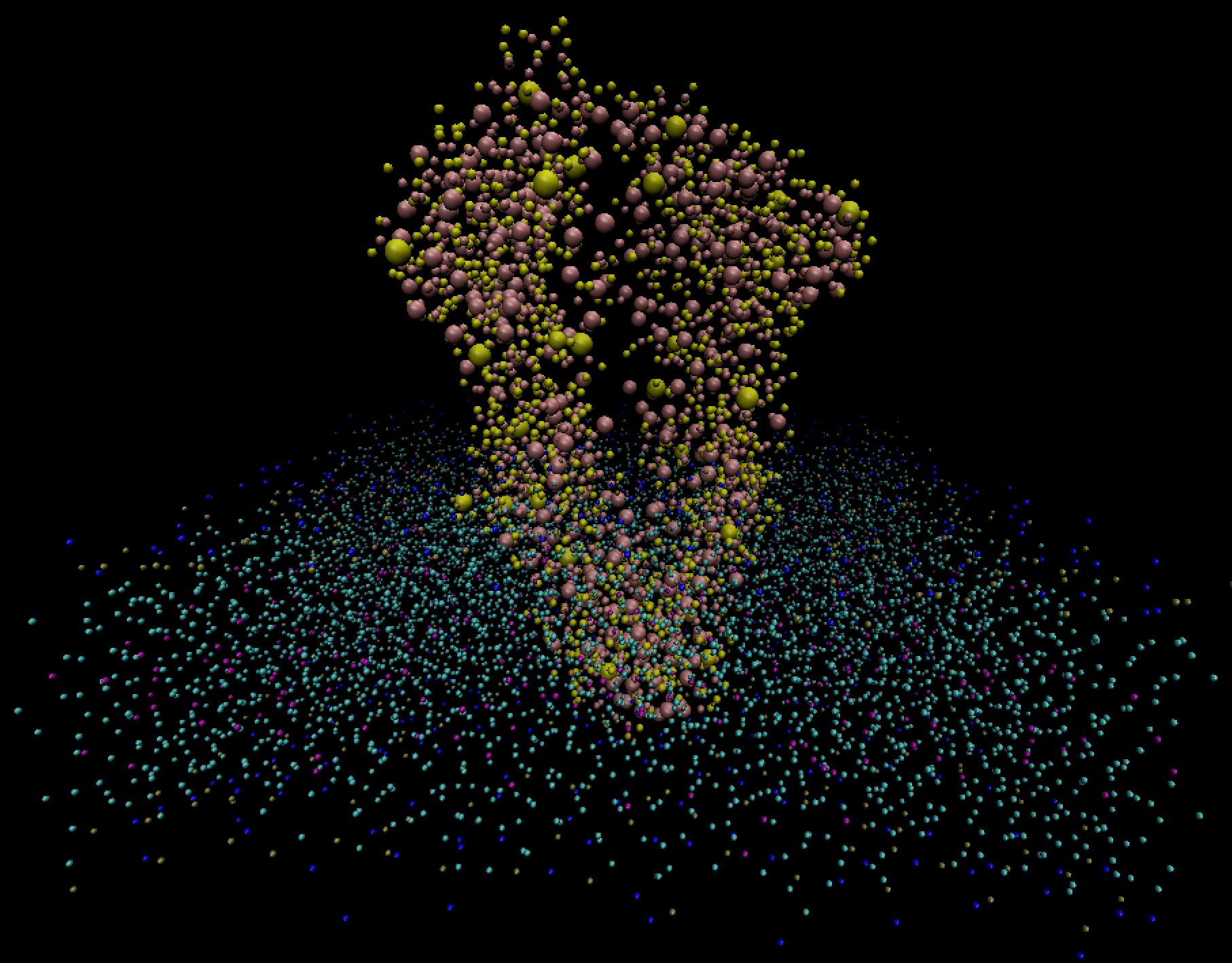
This simulation shows that the P-gp molecule acts as a lipid transporter protein, flipping membrane’s lipids from the inner to the outer leaflet. We asked participants to visualize this process, finding those lipids that bind to the protein.
Linked humanities data
The Orlando British Women’s Writing Dataset (Release 1: Biography and Bibliography) provided a rich set of linked open data representing women’s literary history from the beginnings to the present, concentrated on writing in English in the British Isles but with tentacles out to other languages, literary traditions, and parts of the world. The competition dataset contained 3,321,746 triples stored in the Resource Description Framework (RDF) format which is a standard format for storing linked data called graphs.
To get an idea of the type of information in the dataset, you can browse the Canadian Writing Research Collaboratory (CWRC) ontology’s competency questions. For the purpose of this competition, we suggested to address some more focused questions in the provided dataset.
First place
The first place was taken by Philippe Nazair, a Data Visualization Developer at Université du Québec à Rimouski and part
of the MERIDIAN consortium. Philippe’s visualization is
implemented as a static website with two layers: a graph viewer using the 3d-force-graph web component, and a map viewer
implemented with Leaflet to display markers on the map and written
biographies from Cambridge’s Orlando author’s page found in
the dataset. Below you can see a screen-capture video showing different interactions on this site. All JSON files for
the visualization were created with a Python backend. We especially appreciated sample graphs for radical and liberal
ideologies and other groups, as well as the ability to build more complex graphs by selecting a predicate and setting it
to a certain value, e.g. hasActiveInvolvementIn = socialism, or multiple values; each graph is limited to one
predicate. Individual graph nodes can be displayed as either text or draggable spheres.
(Or click here to watch this video directly on Vimeo.)
Second place
The second prize went to Usman Alim and Roberta Cabral Ramos Mota from the Department of Computer Science at the University of Calgary. Below you can watch one of their visualizations (rendered with OSPRay in ParaView) showing the protein cavity and two highly-interacting PO4 beads with the colour representing time step. This work was selected for their skillful use of Python scripting for the bulk of the analysis: using MDAnalysis library for identifying closely interacting membrane beads, and writing bead positions in VTK to be rendered in ParaView. You can download an edited version of their Python analysis script here.
(Or click here to watch this video directly on Vimeo.)
Third place
The third place was taken by Catherine Winters from the Digital Humanities Innovation Lab at Simon Fraser University. This visualization is powered by D3.js and Three.js, with all JSON data files produced with Python scripting. Not all features of this visualization are complete, so we are not showing it here, but it is likely to remain a side project in the lab over the next few months, with the promise to show clustering based on ideology, religion, or geographic proximity.